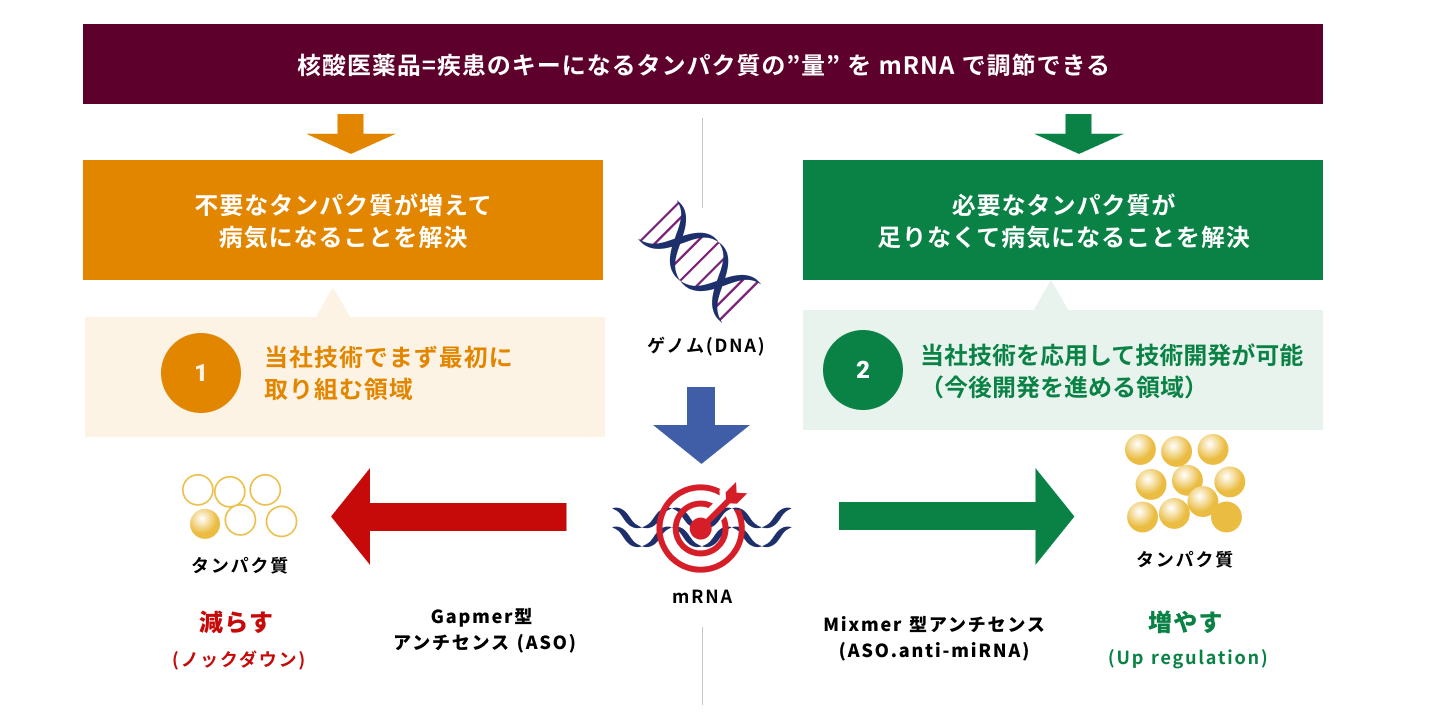
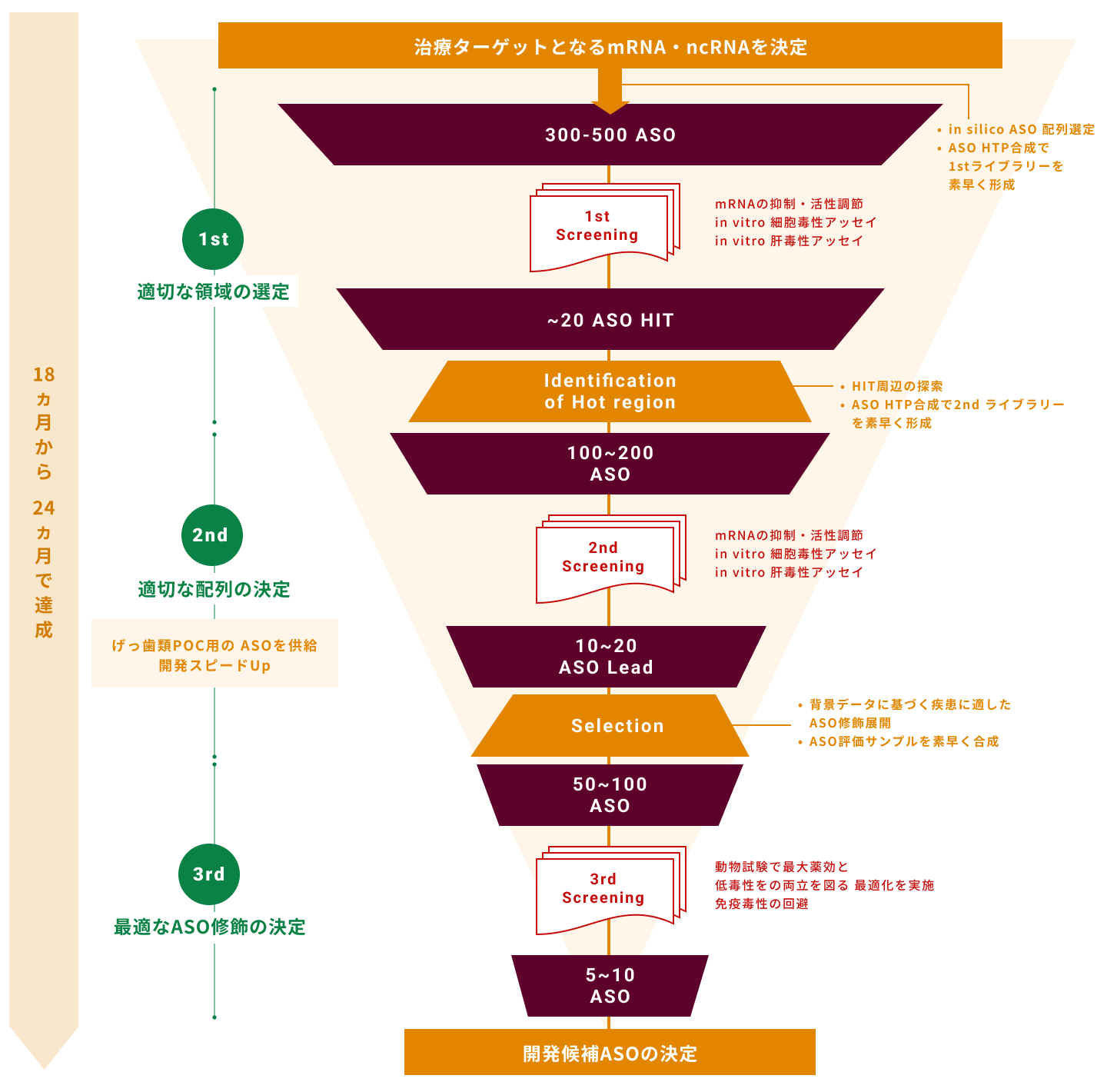
核酸医薬創薬プラットフォーム
当社の創薬プラットフォームは、大阪大学大学院薬学研究科生物有機化学分野(小比賀教授)で蓄積された以下の基盤技術群によって構成されています。
1.修飾核酸群
特徴の異なる4つの修飾核酸があります。
当社の有する核酸群:アンチセンスに最適な素材群

AmNA™
- ヌクレアーゼ耐性の向上
- 塩基特異的なハイブリの向上
- 肝毒性の低減

scpBNA™
- ヌクレアーゼ耐性の向上
- 塩基特異的なハイブリの向上
- 肝毒性の低減
- 疎水度を上げることができる

GuNA™
- ヌクレアーゼ耐性の向上
- 塩基特異的なハイブリの向上
- 鎖中へのプラスチャージ付与
- 神経毒性の低減

5’-CP™
- ホスホロチオエートなしでヌクレアーゼ耐性の向上
- オフターゲット効果の低減
- 毒性低減と薬効持続性の両立に大きく寄与
従来型修飾核酸との比較:当社の修飾核酸群は優れた特性
| 修飾 | タイプ | ハイブリ | ヌクレアーゼ耐性 | 毒性 | モノマー合成難易度 |
|---|---|---|---|---|---|
| S化(PS)DNA | ☓ | ◎ | あり | + | |
| 2′-OMe | 2’位修飾 | △ | △(PS必須) | なし | ++ |
| 2′-F | 2’位修飾 | △ | △(PS必須) | △ | ++ |
| 2′-MOE | 2’位修飾 | ◯ | △(PS必須) | なし | +++ |
| LNA | 架橋型 | ◎ | ◯(PS必須) | あり(肝毒性) | ++++ |
| S-cEt | 架橋型 | ◎ | ◎(部分PS可能) | ◎ | +++++++ |
| AmNA® | 架橋型 | ◎ | ◎(部分PS可能) | ◎ | ++++ |
| GuNA® | 架橋型 | ◎ | ◎(部分PS可能) | ◯(予備情報) | ++++ |
| scpBNA® | 架橋型 | ◎ | ◎(部分PS可能) | ◎(予備情報) | ++++ |
| 5’-CP® | 非架橋型 | 〇 | ◎(PS化不要) | ◎ | +++ |
当社の修飾核酸群
2.アンチセンス毒性低減技術
アンチセンス医薬は、肝毒性・神経毒性に課題がありました。
大阪大学では、新たな修飾核酸と塩基部修飾を用い肝毒性が低減されることを見出しました。
当社では、架橋型修飾核酸と非架橋型核酸を組み合わせた神経毒性低減技術を開発し、毒性の最小化を目指した技術のアンチセンス医薬実装を進めています。
- 1)ギャップマー型アンチセンスにおけるWing領域の構造最適化
- 2)ギャップマー型アンチセンスにおけるGap領域の構造最適化
本研究はAMEDの医療研究開発革新基盤創成事業(CiCLE)に採択されました。
当社は本研究により創出された特許について、包括的なライセンスを大阪大学から受けています。
3.配列設計技術及びアンチセンス創出ワークフロー
アンチセンスを医薬品として開発するにあたり、どの領域に設計するかは重要な要素です。
当社では、大阪大学及び医薬基盤・健康・栄養研究所で培われた基礎技術から、
より候補配列を広く得られるように自社検討を進め、設計技術を立ち上げています。
文献情報
-
AmNA™
Yahara A, Shrestha AR, Yamamoto T, Hari Y, Osawa T, Yamaguchi M, Nishida M, Kodama T, Obika S.
Amido-Bridged Nucleic Acids (AmNAs): Synthesis, Duplex Stability, Nuclease Resistance, and in Vitro Antisense Potency, ChemBioChem, 2012, 13, 2513 – 2516Yamamoto T, Yahara A, Waki R, Yasuhara H, Wada F, Harada-Shiba M, Obika S.
Amido-bridged nucleic acids with small hydrophobic residues enhance hepatic tropism of antisense oligonucleotides in vivo, Org Biomol Chem. 2015,13,12, 3757-65Setoguchi K, Cui L, Hachisuka N, Obchoei S, Shinkai K, Hyodo F, Kato K, Wada F, Yamamoto T, Harada-Shiba M, Obika S, Nakano K.
Antisense Oligonucleotides Targeting Y-Box Binding Protein-1 Inhibit Tumor Angiogenesis by Downregulating Bcl-xL-VEGFR2/-Tie Axes, Molecular Therapy: Nucleic Acids, 2017, 9, 170-181Shimojo M, Kasahara Y, Inoue M, Tsunoda S, Shudo Y, Kurata T, Obika S.
A Gapmer Antisense Oligonucleotide Targeting SRRM4 Is a Novel Therapeutic Medicine for Lung. Cancer, Sci. Rep. 2019, 9,1Uehara T, Choong CJ, Nakamori M, Hayakawa H, Nishiyama K, Kasahara Y, Baba K, Nagata T, Yokota T, Tsuda H, Obika S, Mochizuki H.
Amido-bridged Nucleic Acid (AmNA)-modified Antisense Oligonucleotides Targeting -synuclein As A Novel Therapy For Parkinson’s disease, Scientific Reports, 2019, 9:7567Kanda M, Kasahara Y, Shimizu D, Miwa T, Umeda S, Sawaki K, Nakamura S, Kodera Y, Obika S.
Amido-Bridged Nucleic Acid-Modified Antisense Oligonucleotides Targeting SYT13 to Treat Peritoneal Metastasis of Gastric Cancer, Molecular Therapy: Nucleic Acids, 2020, 22, 791-802Wada F, Yamamoto T, Kobayashi T, Tachibana K, Ito KR, Hamasaki M, Kayaba Y, Terada C, Yamayoshi A, Obika S, Harada-Shiba M.
Drug Discovery and Development Scheme For Liver-targeting Bridged Nucleic Acid Antisense Oligonucleotides, Molecular Therapy: Nucleic Acids, 2021, 26, 957~969Shimo T, Nakatsuji Y, Tachibana K, Obika S.
Design and In Vitro Evaluation of Splice-Switching Oligonucleotides Bearing Locked Nucleic Acids, Amido-Bridged Nucleic Acids, and Guanidine-Bridged Nucleic Acids, Int. J. Mol. Sci. 2021, 22, 3526.Yoshida M, Oda C, Mishima K, Tsuji I, Obika S, Shimojo M.
An Antisense Amido-bridged Nucleic Acid Gapmer Oligonucleotide Targeting SRRM4 Alters REST Splicing And Exhibits Anti-tumor Effects in Small Cell Lung Cancer And Prostate Cancer Cells, Cancer Cell Int, 2023, 23, 8Kanda M, Takano N, Miyauchi H, Ueda K, Mizuno M, Kasahara Y, Kodera Y, Obika S.
Preclinical Toxicological Assessment of Amido‑bridged Nucleic Acid‑modified Antisense Oligonucleotides Targeting Synaptotagmin XIII For Intra‑abdominal Treatment Of Peritoneal Metastasis Of Gastric Cancer, Gastric Cancer , 2024, 27,1229–1241Kuroda M, Kasahara Y, Hirose M, Yamaguma H, Oda M, Nagao C, Mizuguchi K.
Construction of a Tm-value prediction model and molecular dynamics study of AmNA-containing gapmer antisense oligonucleotide, Molecular Therapy: Nucleic Acids, 2024, 35, 3, 102272Uchibori Y, Suekuni M, Kokaji Y, Yoshida K, Kiyono T, Kasahara Y, Fujita M.
AmNA-Modified Antisense Oligonucleotide Targeting MCM8 as a Cancer-Specific Chemosensitizer for Platinum Compounds, Cancer Science, 2025; 116, 1405–1416Kawanobe T, Asano S, Kandori H, Aoki M, Shrestha AR, Sekiguchi K, Yokoyama K, Fukuda R, Umemoto T.
Hepatotoxicity Reduction Profiles of Antisense Oligonucleotides Containing Amido-Bridged Nucleic Acid and 2′-O,4′-C-Spirocyclopropylene Bridged Nucleic Acid, Nucleic Acid Ther, 2025, 35, 3, 114-124Sowa N, Horie T, Ide Y, Baba O, Kora K, Yoshida T, Nakamura Y, Matsumoto S, Matsushita K, Imanaka M, Zou F, Kume E, Kojima H, Qian Q, Kimura K, Otsuka R, Hara N, Yamasaki T, Otani C, Tsujisawa Y, Takaya T, Nishimura C, Watanabe D, Hasegawa K, Kotera J, Oka K, Fujita R, Takemiya A, Sasaki T, Kasahara Y, Obika S, Kimura T, Ono K.
MicroRNA-33 inhibition ameliorates muscular dystrophy by enhancing skeletal muscle regeneration, EMBO Molecular Medicine, 2025, 17, 1902-1925 -
scpBNA™
https://www.glenresearch.com/reports/gr36-21
Yamaguchi T, Horiba M, Obika S.
Synthesis and Properties of 2’-O,4’-C-spirocyclopropylene Bridged Nucleic Acid (scpBNA), an Analogue of 2’,4’- BNA/LNA Bearing a Cyclopropane Ring, Chem. Commun., 2015, 51, 9737Horiba M, Yamaguchi T, Obika S.
Synthesis of scpBNA-mC, -A, and -G Monomers and Evaluation of the Binding Affinities of scpBNA-Modified Oligonucleotides toward Complementary ssRNA and ssDNA, J. Org. Chem. 2016, 81, 22, 11000-11008.Kawanobe T, Asano S, Kandori H, Aoki M, Shrestha AR, Sekiguchi K, Yokoyama K, Fukuda R, Umemoto T.
Hepatotoxicity Reduction Profiles of Antisense Oligonucleotides Containing Amido-Bridged Nucleic Acid and 2′-O,4′-C-Spirocyclopropylene Bridged Nucleic Acid, Nucleic Acid Ther, 2025, 35, 3, 114-124 -
Dual Modification (Base Modification of scpBNA)
Habuchi T, Yamaguchi T, Aoyama H, Horiba M, Ito KR, Obika S.
Hybridization and Mismatch Discrimination Abilities of 2′,4′-Bridged Nucleic Acids Bearing 2‑Thiothymine or 2‑Selenothymine Nucleobase, J. Org. Chem. 2019, 84, 1430-1439.Sakurai Y, Yamaguchi T, Yoshida T, Horiba M, Inoue T, Obika S.
Synthesis and Properties of Nucleobase-Sugar Dual Modified Nucleic Acids: 2′‑OMe-RNA and scpBNA Bearing a 5‑Hydroxycytosine Nucleobase, J. Org. Chem., 2023, 88, 154−162 -
GuNA™
Shrestha AR, Kotobuki Y, Hari Y, Obika S.
Guanidine Bridged Nucleic Acid (GuNA): an Effect of a Cationic Bridged Nucleic Acid on DNA Binding Affinity, Chem Commun 2014, 50, 5, 575-7Horie N, Kumagai S, Kotobuki Y, Yamaguchi T, Obika S.
Facile Synthesis and Fundamental Properties of an N-methylguanidine-bridged Nucleic Acid (GuNA[NMe]), Org Biomol Chem. 2018,16, 35, 6531-6536.Kumagai S, Sawamoto H, Takegawa-Araki T, Arai Y, Yamakoshi S, Yamada K, Ohta Tetsuya, Kawanishi E, Horie N, Yamaguchi T, Obika S.
Synthesis and Properties of GuNA purine/pyrimidine Nucleosides and Oligonucleotides, Org. Biomol. Chem., 2020, 18, 9461Horie N, Yamaguchi T, Kumagai S, Obika S.
Synthesis and properties of oligonucleotides modified with an N-methylguanidine-bridged nucleic acid (GuNA[Me]) bearing adenine, guanine, or 5-methylcytosine nucleobases, Beilstein J. Org. Chem. 2021, 17, 622–629.Shimo T, Nakatsuji Y, Tachibana K, Obika S.
Design and In Vitro Evaluation of Splice-Switching Oligonucleotides Bearing Locked Nucleic Acids, Amido-Bridged Nucleic Acids, and Guanidine-Bridged Nucleic Acids, Int. J. Mol. Sci. 2021, 22, 3526.Takegawa-Araki T, Kumagai S. Yasukawa K, Kuroda M, Sasaki T, Obika S.
Structure−Activity Relationships of Anti-microRNA Oligonucleotides Containing Cationic Guanidine-Modified Nucleic Acids, J. Med. Chem. 2022, 65, 2139−2148Sasaki T, Hirakawa Y, Yamairi F, Kurita T, Murahashi K, Nishimura H, Iwazaki N, Yasuhara H, Tateoka T, Ohta T, Obika S, Kotera Jun.
Altered Biodistribution and Hepatic Safety Profile of a Gapmer Antisense Oligonucleotide Bearing Guanidine-Bridged Nucleic Acids, Nucleic Acid Ther, 2022, 32, 3, 177-184Yamaguchi T, Horie N, Aoyama H, Kumagai S, Obika S.
Mechanism of the Extremely High Duplex-forming Ability of Oligonucleotides Modified With N-tert-butylguanidine- or N-tert-butyl-N′-methylguanidine-bridged Nucleic Acids Nucleic acid research, 2023, 51, 7749 -
5′-CP™
Kuroda T, Yoshioka K, Mon SSL, Katsuyama M, Sato K, Isogai E, Yoshida-Tanaka K, Iwata-Hara R, Yamaguchi T, Obika S, Yokota T.
Unraveling and controlling late-onset neurotoxicity of antisense oligonucleotides through strategic chemical modifications, Molecular Therapy: Nucleic Acids , 2025, September 12, 102692