Platform of oligonucleotide therapeutics discovery
Our drug discovery platform consists of following basic technologies accumulated in Professor Obika’s laboratory (Osaka University Graduate School of Pharmaceutical Sciences, Bioorganic Chemistry).
1. Modified nucleic acids
We offer four modified nucleic acids, comprising three bridged nucleic acid analogs and one structurally enhancedDNA analog
Modified XNAs: Superior Platforms for Antisense Applications

AmNA™
- Improvement of nuclease resistance
- Improvement of base-specific hybridization
- Reduction in hepatotoxicity

scpBNA™
- Improvement of nuclease resistance
- Improvement of base-specific hybridization
- Reduction in hepatotoxicity
- Adds hydrophobicity in the sequence

GuNA™
- Improvement of nuclease resistance
- Improvement of base-specific hybridization
- Apply a positive charge to the chain
- Reduction in immunotoxicity

5′-CP™
- Improvement of nuclease resistance without phosphorothioation
- Reduction in off-target effects
- Reduction in hepato- and neurotoxicity
Comparison with other modifications: Our modification group endows your oligonucleotides with superior properties.
| Modification | Type | Hybridize | Nuclease resistance | Hepatotoxicity | Mfg. Cost |
|---|---|---|---|---|---|
| PS DNA | + | +++++ | ++ | + | |
| 2′-OMe | 2′-modify | ++ | + | – | ++ |
| 2′-F | 2′-modify | ++ | + | ++ | ++ |
| 2′-MOE | 2′-modify | ++ | ++ | + | +++ |
| LNA | Bi-cyclic | ++++ | ++ | +++++ (Hepatotoxicity) |
++++ |
| S-cEt | Bi-cyclic | ++++ | +++ | + | +++++++ |
| AmNA™ | Bi-cyclic | ++++ | +++ | + | +++ |
| GuNA™ | Bi-cyclic | ++++ | ++++ | + | ++++ |
| scpBNA™ | Bi-cyclic | ++++ | +++++ | + | ++++ |
| 5′-CP™ | non-bridge | ++ | ++++ | + (neurotoxicity+) |
++ |
Our modification
2. Antisense toxicity reduction technology
Antisense drugs are associated with the risk of hepatotoxicity.
At Osaka University, they discovered that the following technologies reduce hepatotoxicity by utilizing a new bridged nucleic acid and/or base modification incorporated into the oligonucleotide, and furthermore, they are developing technologies aiming at minimizing hepatotoxicity.
- 1)Base modification of gap segment in gapmer antisense oligonucleotide
- 2)Dual modification of wing segment in gapmer antisense oligonucleotide
This research was selected for AMED’s Cyclic Innovation for Clinical Empowerment (CiCLE) program.
Our company has obtained a comprehensive license from Osaka University for the patents generated through this research.
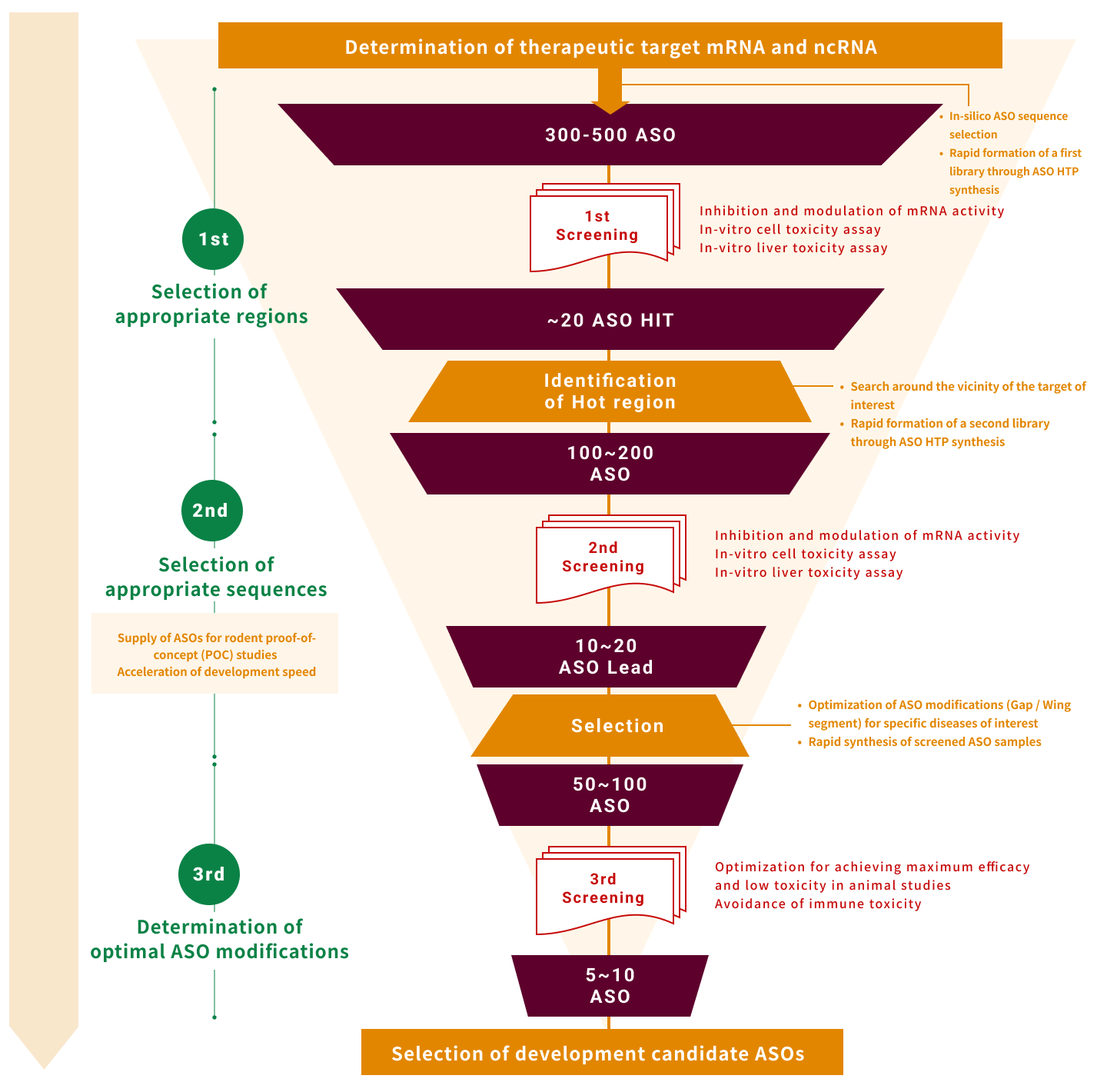
3. Sequence design system
It is important in the design process to decide in which mRNA area to develop antisense therapy.
Based on basic technologies accumulated in Osaka University and NIBIOHN, we are advancing our own research and setting up design technologies to obtain as many candidate sequences as we can.
Literature
-
AmNA™
Yahara A, Shrestha AR, Yamamoto T, Hari Y, Osawa T, Yamaguchi M, Nishida M, Kodama T, Obika S.
Amido-Bridged Nucleic Acids (AmNAs): Synthesis, Duplex Stability, Nuclease Resistance, and in Vitro Antisense Potency, ChemBioChem, 2012, 13, 2513 – 2516Yamamoto T, Yahara A, Waki R, Yasuhara H, Wada F, Harada-Shiba M, Obika S.
Amido-bridged nucleic acids with small hydrophobic residues enhance hepatic tropism of antisense oligonucleotides in vivo, Org Biomol Chem. 2015,13,12, 3757-65Setoguchi K, Cui L, Hachisuka N, Obchoei S, Shinkai K, Hyodo F, Kato K, Wada F, Yamamoto T, Harada-Shiba M, Obika S, Nakano K.
Antisense Oligonucleotides Targeting Y-Box Binding Protein-1 Inhibit Tumor Angiogenesis by Downregulating Bcl-xL-VEGFR2/-Tie Axes, Molecular Therapy: Nucleic Acids, 2017, 9, 170-181Shimojo M, Kasahara Y, Inoue M, Tsunoda S, Shudo Y, Kurata T, Obika S.
A Gapmer Antisense Oligonucleotide Targeting SRRM4 Is a Novel Therapeutic Medicine for Lung. Cancer, Sci. Rep. 2019, 9,1Uehara T, Choong CJ, Nakamori M, Hayakawa H, Nishiyama K, Kasahara Y, Baba K, Nagata T, Yokota T, Tsuda H, Obika S, Mochizuki H.
Amido-bridged Nucleic Acid (AmNA)-modified Antisense Oligonucleotides Targeting -synuclein As A Novel Therapy For Parkinson’s disease, Scientific Reports, 2019, 9:7567Kanda M, Kasahara Y, Shimizu D, Miwa T, Umeda S, Sawaki K, Nakamura S, Kodera Y, Obika S.
Amido-Bridged Nucleic Acid-Modified Antisense Oligonucleotides Targeting SYT13 to Treat Peritoneal Metastasis of Gastric Cancer, Molecular Therapy: Nucleic Acids, 2020, 22, 791-802Wada F, Yamamoto T, Kobayashi T, Tachibana K, Ito KR, Hamasaki M, Kayaba Y, Terada C, Yamayoshi A, Obika S, Harada-Shiba M.
Drug Discovery and Development Scheme For Liver-targeting Bridged Nucleic Acid Antisense Oligonucleotides, Molecular Therapy: Nucleic Acids, 2021, 26, 957~969Shimo T, Nakatsuji Y, Tachibana K, Obika S.
Design and In Vitro Evaluation of Splice-Switching Oligonucleotides Bearing Locked Nucleic Acids, Amido-Bridged Nucleic Acids, and Guanidine-Bridged Nucleic Acids, Int. J. Mol. Sci. 2021, 22, 3526.Yoshida M, Oda C, Mishima K, Tsuji I, Obika S, Shimojo M.
An Antisense Amido-bridged Nucleic Acid Gapmer Oligonucleotide Targeting SRRM4 Alters REST Splicing And Exhibits Anti-tumor Effects in Small Cell Lung Cancer And Prostate Cancer Cells, Cancer Cell Int, 2023, 23, 8Kanda M, Takano N, Miyauchi H, Ueda K, Mizuno M, Kasahara Y, Kodera Y, Obika S.
Preclinical Toxicological Assessment of Amido‑bridged Nucleic Acid‑modified Antisense Oligonucleotides Targeting Synaptotagmin XIII For Intra‑abdominal Treatment Of Peritoneal Metastasis Of Gastric Cancer, Gastric Cancer , 2024, 27,1229–1241Kuroda M, Kasahara Y, Hirose M, Yamaguma H, Oda M, Nagao C, Mizuguchi K.
Construction of a Tm-value prediction model and molecular dynamics study of AmNA-containing gapmer antisense oligonucleotide, Molecular Therapy: Nucleic Acids, 2024, 35, 3, 102272Uchibori Y, Suekuni M, Kokaji Y, Yoshida K, Kiyono T, Kasahara Y, Fujita M.
AmNA-Modified Antisense Oligonucleotide Targeting MCM8 as a Cancer-Specific Chemosensitizer for Platinum Compounds, Cancer Science, 2025; 116, 1405–1416Kawanobe T, Asano S, Kandori H, Aoki M, Shrestha AR, Sekiguchi K, Yokoyama K, Fukuda R, Umemoto T.
Hepatotoxicity Reduction Profiles of Antisense Oligonucleotides Containing Amido-Bridged Nucleic Acid and 2′-O,4′-C-Spirocyclopropylene Bridged Nucleic Acid, Nucleic Acid Ther, 2025, 35, 3, 114-124Sowa N, Horie T, Ide Y, Baba O, Kora K, Yoshida T, Nakamura Y, Matsumoto S, Matsushita K, Imanaka M, Zou F, Kume E, Kojima H, Qian Q, Kimura K, Otsuka R, Hara N, Yamasaki T, Otani C, Tsujisawa Y, Takaya T, Nishimura C, Watanabe D, Hasegawa K, Kotera J, Oka K, Fujita R, Takemiya A, Sasaki T, Kasahara Y, Obika S, Kimura T, Ono K.
MicroRNA-33 inhibition ameliorates muscular dystrophy by enhancing skeletal muscle regeneration, EMBO Molecular Medicine, 2025, 17, 1902-1925 -
scpBNA™
https://www.glenresearch.com/reports/gr36-21
Yamaguchi T, Horiba M, Obika S.
Synthesis and Properties of 2’-O,4’-C-spirocyclopropylene Bridged Nucleic Acid (scpBNA), an Analogue of 2’,4’- BNA/LNA Bearing a Cyclopropane Ring, Chem. Commun., 2015, 51, 9737Horiba M, Yamaguchi T, Obika S.
Synthesis of scpBNA-mC, -A, and -G Monomers and Evaluation of the Binding Affinities of scpBNA-Modified Oligonucleotides toward Complementary ssRNA and ssDNA, J. Org. Chem. 2016, 81, 22, 11000-11008.Kawanobe T, Asano S, Kandori H, Aoki M, Shrestha AR, Sekiguchi K, Yokoyama K, Fukuda R, Umemoto T.
Hepatotoxicity Reduction Profiles of Antisense Oligonucleotides Containing Amido-Bridged Nucleic Acid and 2′-O,4′-C-Spirocyclopropylene Bridged Nucleic Acid, Nucleic Acid Ther, 2025, 35, 3, 114-124 -
Dual Modification (Base Modification of scpBNA)
Habuchi T, Yamaguchi T, Aoyama H, Horiba M, Ito KR, Obika S.
Hybridization and Mismatch Discrimination Abilities of 2′,4′-Bridged Nucleic Acids Bearing 2‑Thiothymine or 2‑Selenothymine Nucleobase, J. Org. Chem. 2019, 84, 1430-1439.Sakurai Y, Yamaguchi T, Yoshida T, Horiba M, Inoue T, Obika S.
Synthesis and Properties of Nucleobase-Sugar Dual Modified Nucleic Acids: 2′‑OMe-RNA and scpBNA Bearing a 5‑Hydroxycytosine Nucleobase, J. Org. Chem., 2023, 88, 154−162 -
GuNA™
Shrestha AR, Kotobuki Y, Hari Y, Obika S.
Guanidine Bridged Nucleic Acid (GuNA): an Effect of a Cationic Bridged Nucleic Acid on DNA Binding Affinity, Chem Commun 2014, 50, 5, 575-7Horie N, Kumagai S, Kotobuki Y, Yamaguchi T, Obika S.
Facile Synthesis and Fundamental Properties of an N-methylguanidine-bridged Nucleic Acid (GuNA[NMe]), Org Biomol Chem. 2018,16, 35, 6531-6536.Kumagai S, Sawamoto H, Takegawa-Araki T, Arai Y, Yamakoshi S, Yamada K, Ohta Tetsuya, Kawanishi E, Horie N, Yamaguchi T, Obika S.
Synthesis and Properties of GuNA purine/pyrimidine Nucleosides and Oligonucleotides, Org. Biomol. Chem., 2020, 18, 9461Horie N, Yamaguchi T, Kumagai S, Obika S.
Synthesis and properties of oligonucleotides modified with an N-methylguanidine-bridged nucleic acid (GuNA[Me]) bearing adenine, guanine, or 5-methylcytosine nucleobases, Beilstein J. Org. Chem. 2021, 17, 622–629.Shimo T, Nakatsuji Y, Tachibana K, Obika S.
Design and In Vitro Evaluation of Splice-Switching Oligonucleotides Bearing Locked Nucleic Acids, Amido-Bridged Nucleic Acids, and Guanidine-Bridged Nucleic Acids, Int. J. Mol. Sci. 2021, 22, 3526.Takegawa-Araki T, Kumagai S. Yasukawa K, Kuroda M, Sasaki T, Obika S.
Structure−Activity Relationships of Anti-microRNA Oligonucleotides Containing Cationic Guanidine-Modified Nucleic Acids, J. Med. Chem. 2022, 65, 2139−2148Sasaki T, Hirakawa Y, Yamairi F, Kurita T, Murahashi K, Nishimura H, Iwazaki N, Yasuhara H, Tateoka T, Ohta T, Obika S, Kotera Jun.
Altered Biodistribution and Hepatic Safety Profile of a Gapmer Antisense Oligonucleotide Bearing Guanidine-Bridged Nucleic Acids, Nucleic Acid Ther, 2022, 32, 3, 177-184Yamaguchi T, Horie N, Aoyama H, Kumagai S, Obika S.
Mechanism of the Extremely High Duplex-forming Ability of Oligonucleotides Modified With N-tert-butylguanidine- or N-tert-butyl-N′-methylguanidine-bridged Nucleic Acids Nucleic acid research, 2023, 51, 7749 -
5′-CP™
Kuroda T, Yoshioka K, Mon SSL, Katsuyama M, Sato K, Isogai E, Yoshida-Tanaka K, Iwata-Hara R, Yamaguchi T, Obika S, Yokota T.
Unraveling and controlling late-onset neurotoxicity of antisense oligonucleotides through strategic chemical modifications, Molecular Therapy: Nucleic Acids , 2025, September 12, 102692